Russian 23andme format 9 files
In this video, we will explore the genetics of Central Russians. I gathered 9 genomes from the Reich Lab’s Human Origins dataset and converted the samples into microarray format.
3 samples are from the town of Kashin in Tver oblast, 3 samples are from Mologa in Yaroslavl Oblast, and additional 3 samples are from the town of Bolkhov in Oryol oblast. Then, I ran those samples through my Trait Predictor tool for DNA analysis.
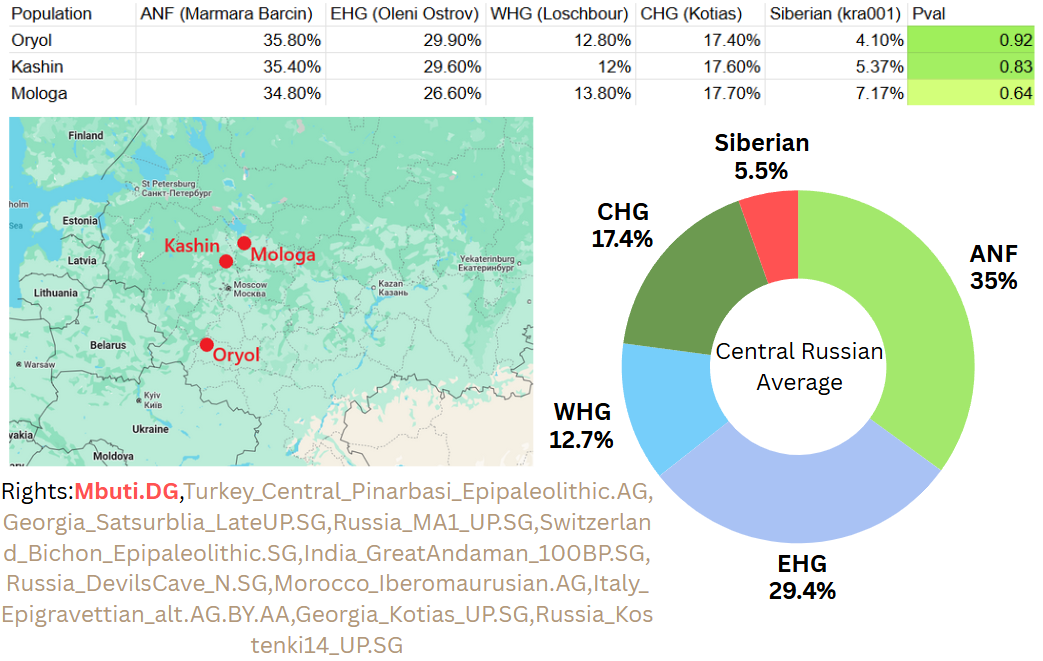
qpAdm analysis of these russians reveals that they are similar to other Europeans genetically. Just as in other Europeans, the dominant ancestry component in these central Russians is Anatolian Neolithic farmer. Here are the averages of 3 models I’ve made for these Central Russians using Kashin, Oryol, and Mologa samples as targets.
And here are the results of G25 analysis of these samples. The three Kashin samples are closest to various southern and central russians, but Ukrainians show up among the closest populations as well. The Mologa samples appear to be more northern shifted, clustering with western Russians, Balts, and Belarusians, and further from Ukrainians. The Oryol samples, on the other hand, appear to be very southern shifted, clustering closest to Northern Ukrainians. Keep in mind these G25 coordinates are simulated by me using the Eurogenes K36 calculator.
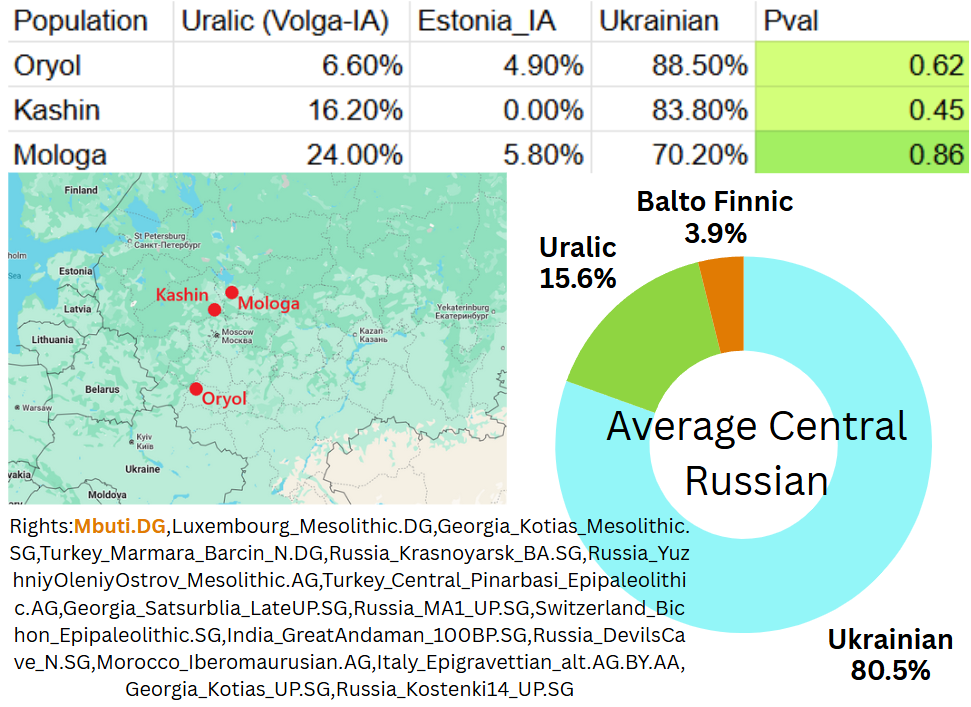
qpAdm analysis of these central russians reveals they are of mostly Slavic origin with just minor Uralic contributions. Out of these central Russian populations, the Mologa samples are the least Slavic and the Oryol samples most slavic, with the Kashin samples occupying an intermediate position.